If you have seen me give a talk in the last couple of years, chances are you would have heard a bit about Shigella sonnei. This is because it has been my favourite project in recent years, for two main reasons:
(1) it involved looking in-depth at phylogeography and evolution of the same organism at two different scales – first globally, over hundreds of years and then locally in Vietnam, over about 15 years; and
(2) it was done with two people I really enjoy working with – Steve Baker (based at the Oxford University Clinical Research Unit in Vietnam) and Nick Thomson (based at the Sanger Institute).
Here are the papers:
Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe
Holt KE, et al. Nature Genetics 2012 [PubMed]
This study used whole genome sequencing of a global collection of 132 Shigella sonnei, an increasingly important cause of dysentery, to reconstruct the evolutionary history of the bacterium. Phylogenetic analysis showed that the current S. sonnei population descends from a common ancestor that existed less than 500 years ago and that diversified into several distinct lineages with unique characteristics. Furthermore the analysis suggests that the majority of this diversification occurred in Europe and was followed by more recent establishment of local pathogen populations on other continents, predominantly due to the pandemic spread of a single, rapidly evolving, multidrug-resistant lineage.
Commentaries on the paper are available in Nature Genetics and Nature Reviews Gastroenterology and Hepatology.
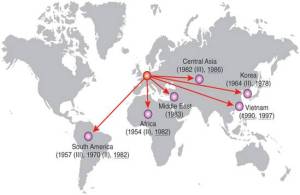
Dissemination of S. sonnei lineages out of Europe. Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics 44:1056, copyright 2012.
Tracking the establishment of local endemic populations of an emergent enteric pathogen
Holt KE, et al. PNAS 2013 [PubMed]
This study continues the Shigella sonnei story by examining the arrival of the rapidly evolving multidrug-resistant lineage in one particular country – Vietnam. We sequenced over 250 genomes of S. sonneiisolated over a 15-year period, and found that the multidrug-resistant lineage successfully established itself in Ho Chi Minh City, pushing out other dysentery-causing bacteria to become the dominant cause of dysentery.
This was likely helped by the acquisition of a colicin (toxin) system that enabled it to kill competing bacteria it came into contact with (including otherShigella), forming a new clone we called the VN (Viet Nam) clone. The VN clone spread to other cities in Vietnam, and we found evidence of convergent evolution of drug resistance mutations and plasmids in all three local populations we examined.
